AI Model and Data Verification Methodology
Our Quality Assurance (QA) framework is a comprehensive system designed to ensure the accuracy, reliability, and consistency of the clinical data we provide. This document provides a deep dive into our quality process, which is built on a fundamental principle: first, we verify the AI; then, we validate the data.
This three-step methodology combines physician-led review, statistical validation, and continuous monitoring to ensure quality at every stage.
- Verification: Confirming the accuracy of AI-generated data against the original source. This ensures our models are performing correctly.
- Validation: Assessing the overall quality, conformance, and plausibility of the final, OMOP-formatted dataset using a standardized, objective framework.
- Final Clinical Plausibility Review: A final, holistic review of the dataset by a clinical expert to ensure it is fit for the specific research purpose.
1. Verification
Verification is a physician-led process to ensure that the data extracted and normalized by our AI is correct before it is integrated into the OMOP CDM. This process is performed at different levels of granularity to ensure comprehensive accuracy:
-
Instance-level Verification: At the most detailed level, physicians verify individual data points as they appear in the clinical notes. For a specific concept, they confirm that the extraction is accurate and the context is correct. For instance, the system is trained to distinguish between a patient’s actual diagnosis (“patient has hypertension”), a negated statement (“patient denies hypertension”), or a family history (“mother has hypertension”).
The Remote Source Data Verification (rSDV), a process where source data from study sites is verified remotely by a centralized specialist team rather than on-site at each location, explicitly confirms that these nuances are captured correctly.
-
Patient-level Verification: This is a broader check to confirm that a patient’s overall profile, based on all the extracted data, is clinically coherent. This is particularly important for cohort-based studies, where physicians verify that a patient correctly belongs to a specific subcohort.
This multi-faceted approach ensures that our verification is robust, from the smallest data point to the overall patient profile. The core of this process involves several key steps, with a sample size that is scientifically determined for each hospital, cohort, and clinical concept using a Bayesian statistical framework. Our methodology calculates the minimum sample size required to achieve a 95% level of confidence with a 5% margin of error, ensuring our performance metrics are statistically robust. The verification process continues until this pre-defined level of precision is achieved.
Remote Source Data Verification (rSDV)
Our physicians perform rSDV) on a significant sample of the data. They are presented with the original clinical text alongside the AI’s output and must assess whether the AI’s inference is accurate. This process evaluates:
- True Positives (TP): Instances where the AI correctly identified and coded a piece of clinical information.
- False Positives (FP): Instances where the AI incorrectly identified or coded information.
Each annotation is cross-verified by at least two physicians to ensure consistency.

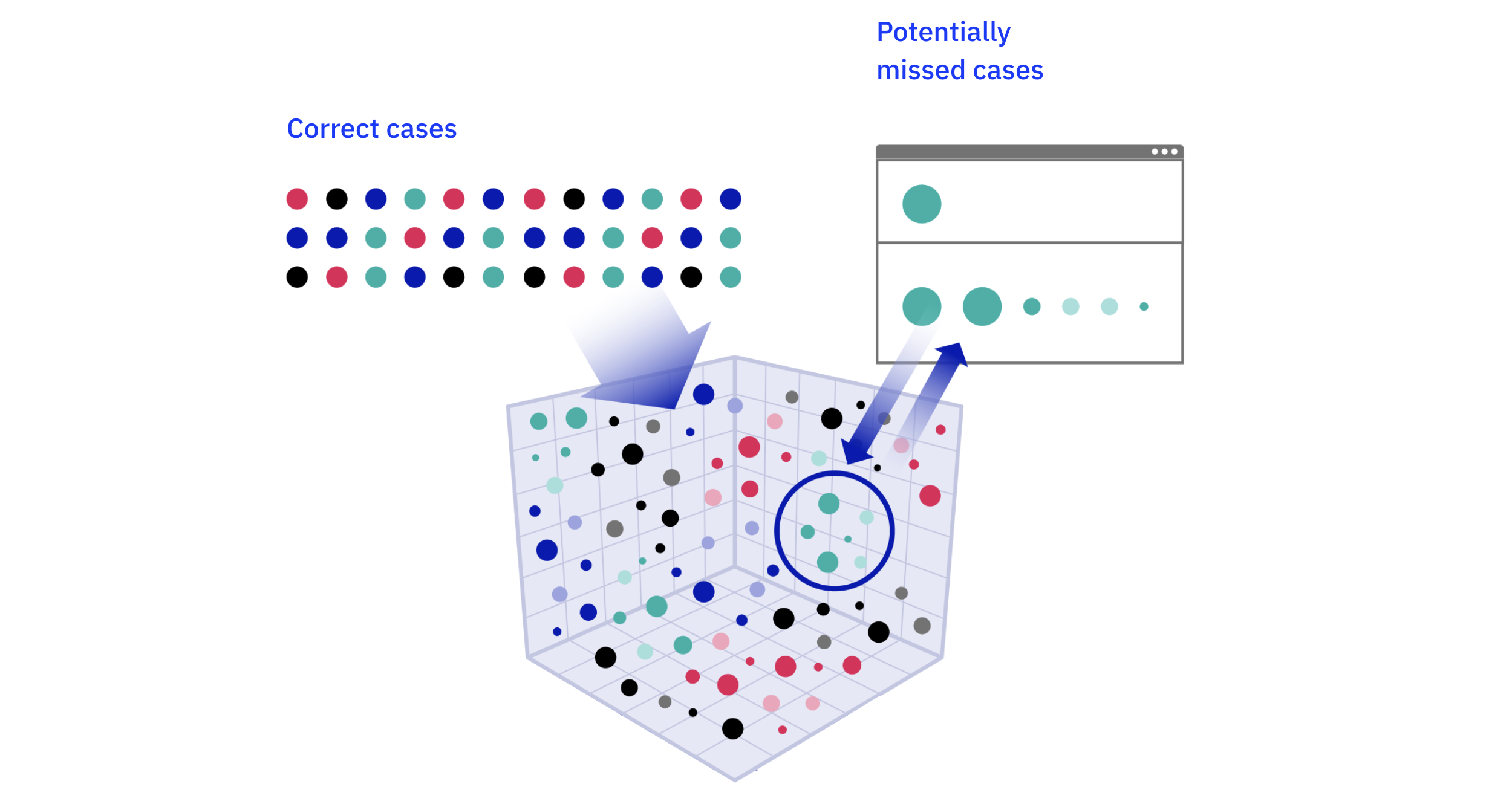
False Negative (FN) Detection
False negatives—relevant clinical information that the AI missed—are difficult to find by random sampling. To address this, we use a semantic similarity search internally develelped by us1. The system identifies data points that have been correctly identified and then searches the entire dataset for similar, but previously unrecognized, instances. The performance of our semantic similarity tool is regularly benchmarked against manual physician review to ensure its accuracy in identifying potential false negatives. These potential false negatives are then sent for rSDV to confirm if they are true missed cases.
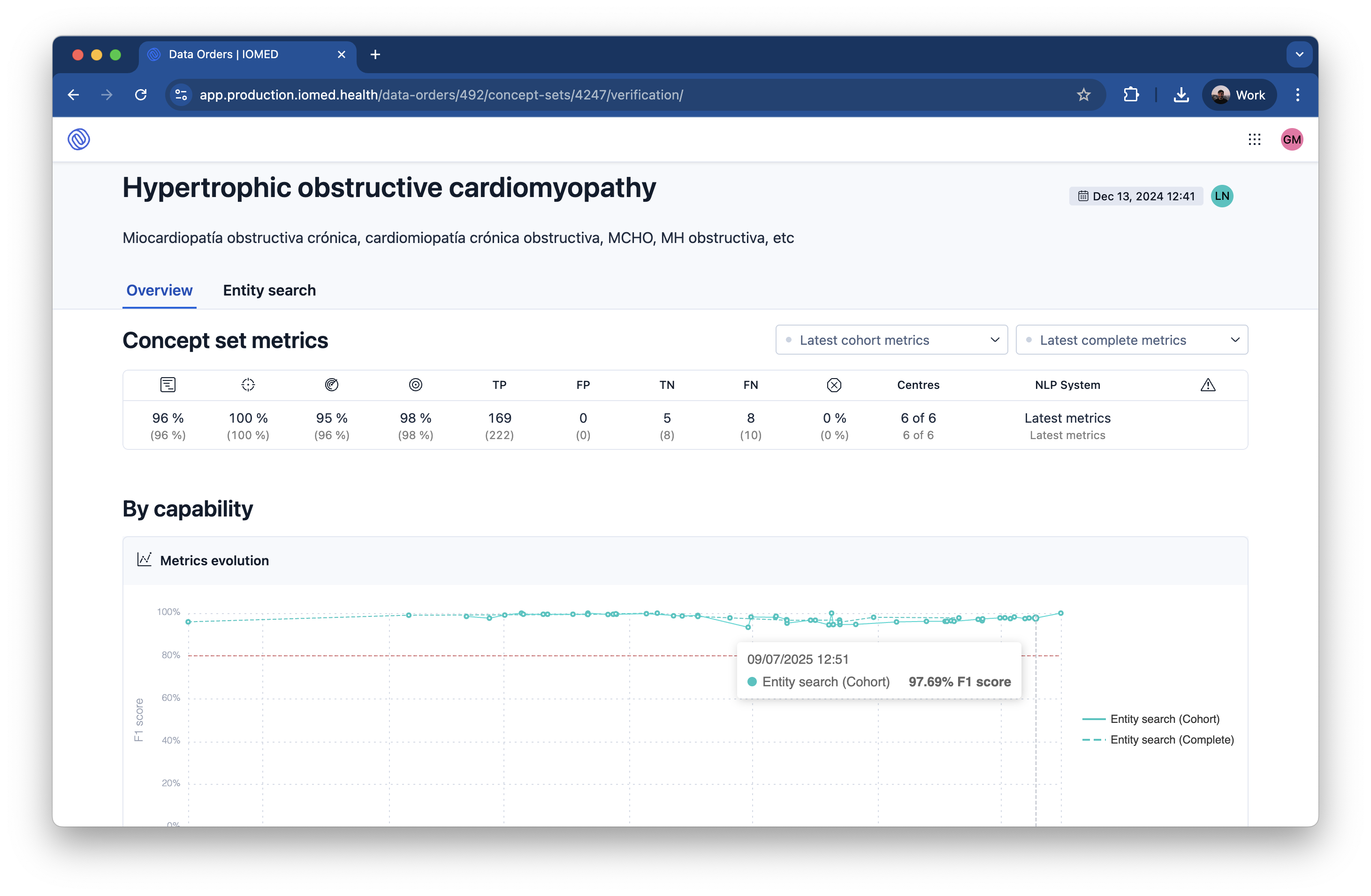
Performance Metrics
We measure the effectiveness of our AI models using standard performance metrics derived from the verification process. Our AI models are continuously monitored and retrained to achieve target performance metrics of >95% Precision and >90% Recall for critical clinical concepts.
- Precision: The proportion of AI-generated data that is correct.
- Recall: The proportion of all relevant data that the AI successfully extracted.
- F1-Score: The harmonic mean of Precision and Recall, providing a single measure of a model’s accuracy.
Annotator Evaluation: Monitoring Our Experts
The quality of our AI is directly related to the quality of the expert annotations used to train and verify it. We use a real-time algorithm to monitor the performance of our physician annotators.
- Inter-Annotator Agreement: We use Fleiss’ Kappa to measure the consistency of annotations among different physicians. Our annotators consistently achieve a Fleiss’ Kappa of 0.81, indicating substantial agreement.
- Golden Annotators: A group of our most experienced physicians, or “golden annotators,” continuously annotate data samples. The performance of all other annotators is regularly compared against this benchmark to ensure they meet the required standards. Our annotators average a 91% agreement score with the golden annotators.
- Time per Task (TPT): We monitor the time it takes for annotators to complete tasks to ensure efficiency and engagement. The average TPT is 48 seconds, which has remained stable over time.
2. Systematic Data Validation
While Verification ensures the accuracy of our AI extraction, Validation ensures the overall quality of the final OMOP dataset. This stage is not a qualitative review; it is a systematic, software-driven process based on the standardized OHDSI Data Quality Dashboard (DQD) framework.
This framework is built upon the Kahn Framework for Data Quality2, which organizes checks into three key dimensions:
- Completeness: We measure whether all necessary data elements are present. The DQD includes checks like
measureValueCompletenessto ensure all required fields are populated. Our AI-driven process has been shown to increase the volume of unique data points by 40% and the diversity of clinical concepts by over 65% compared to structured data alone. - Conformance: We verify that data adheres to the expected format and structure of the OMOP CDM. This includes checks like
cdmDatatype(verifying correct data types) andisForeignKey(ensuring relational integrity between tables). - Plausibility: We run automated checks to ensure the data is believable and logically consistent. This includes temporal checks (e.g.,
plausibleStartBeforeEndto ensure start dates are before end dates) and clinical logic checks (e.g.,plausibleBeforeDeathto ensure events do not occur long after a patient’s death).
This systematic validation provides an objective, reproducible, and transparent assessment of the final dataset’s quality, ensuring it is robust and ready for analysis.
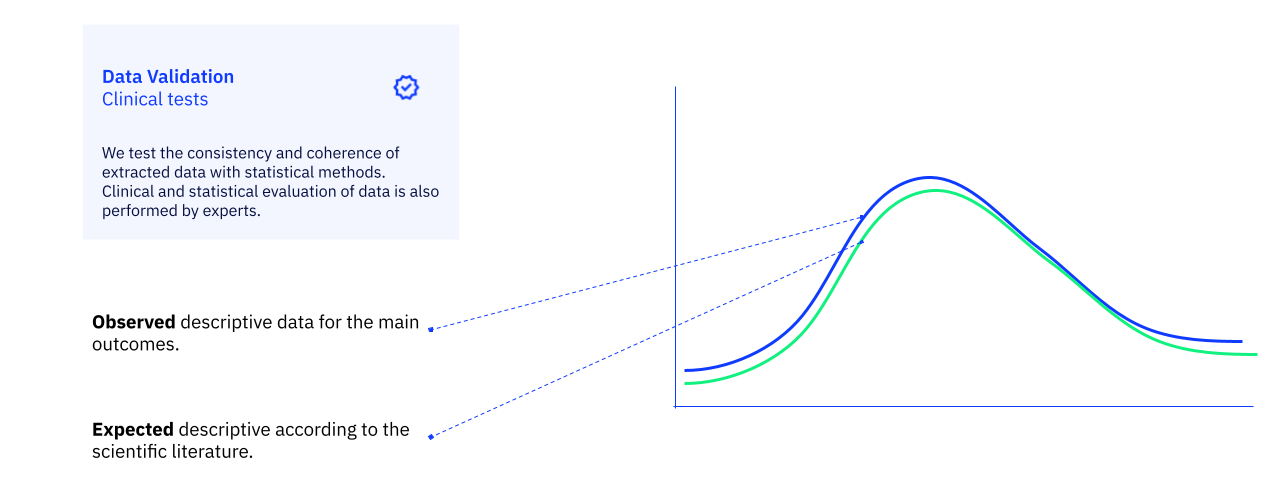
3. Final Clinical Plausibility Review
After the automated validation checks are complete, a physician on our clinical team performs a final, holistic review of the dataset. This is not a check of individual data points, but rather a qualitative assessment of the complete cohort-level data.
The goal is to ensure that the final dataset is clinically coherent and appropriate for the specific research question. The reviewing physician assesses the overall patient profiles, examines the distribution of key characteristics, and uses their clinical expertise to confirm that the data “makes sense.” This final, human-expert sign-off ensures that the dataset is not just technically correct, but also clinically valid and fit-for-purpose.
-
Maria Quijada, Maria Vivó, Álvaro Abella-Bascarán, Paula Chocrón, and Gabriel de Maeztu. 2022. A Framework for False Negative Detection in NER/NEL. In Natural Language Processing and Information Systems: 27th International Conference on Applications of Natural Language to Information Systems, NLDB 2022, Valencia, Spain, June 15–17, 2022, Proceedings. Springer-Verlag, Berlin, Heidelberg, 323–330. https://doi.org/10.1007/978-3-031-08473-7_30 ↩
-
Kahn, M. G., Callahan, T. J., Barnard, J., Bauck, A. E., Brown, J., Davidson, B. N., Estiri, H., Goerg, C., Holve, E., Johnson, S. G., & Liaw, S.-T. (2016). A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data. In EGEMS (Vol. 4, Issue 1, p. 1244). Ubiquity Press. https://doi.org/10.13063/2327-9214.1244 ↩