Patient-Level Characterisation Guide
Introduction & Purpose
A Patient-Level Characterisation study is the observational research equivalent of creating a “Table 1” in a clinical trial. Its primary purpose is to generate a detailed clinical and demographic profile of a specific group of patients (a cohort). This answers the fundamental question: “Who are these patients?”
This type of analysis is a cornerstone of transparent and reproducible research. By providing a comprehensive baseline description of the study population, it helps researchers and readers understand the context of the study, assess the generalisability of the findings, and identify potential sources of confounding or bias.
Study Design
The design is a descriptive cohort analysis. It focuses on summarising the characteristics of one or more cohorts of patients at a specific point in time (the index date), or within a defined time window before it.
Participants
The study includes one or more cohorts of interest. These cohorts are typically defined by a shared characteristic, such as:
- A new diagnosis of a specific condition.
- The initiation of a particular medication.
- Undergoing a specific medical procedure.
A key requirement is that participants have a period of data visibility (e.g., one year) before their index date to allow for the assessment of baseline characteristics.
Exposures / Covariates
In this context, there is no “exposure” in the comparative sense. Instead, the analysis focuses on summarising a wide range of patient covariates (characteristics) present at or before the index date. These can include:
- Demographics: Age, gender, race, ethnicity.
- Clinical History: All recorded medical conditions, often summarised into comorbidity scores like the Charlson Comorbidity Index.
- Medication History: All prior medications the patients have used.
- Procedures: A history of medical procedures.
Outcomes
The “outcomes” of this study are the summary statistics for the covariates of interest. The analysis produces tables and visualisations that describe the distribution of these characteristics within the cohort. This can include:
- For categorical variables: Frequencies and percentages (e.g., % of patients with a history of diabetes).
- For continuous variables: Means, medians, and standard deviations (e.g., mean age of the cohort).
Follow-up
There is typically no follow-up period after the index date in a pure characterisation study. The focus is entirely on the baseline period before the index date.
Analyses
The analysis is descriptive and involves summarising the covariates. This can be done in two ways:
- Large-Scale Characterisation: An automated process that summarises thousands of clinical features from the database to provide an unbiased, data-driven overview of the cohort.
- Pre-Specified Characterisation: An analysis focused on a limited set of clinically important covariates that have been defined in advance by the researchers.
The results are typically presented in a summary table, often referred to as “Table 1,” which provides a comprehensive snapshot of the cohort.
How to Implement This Study
Let’s implements the study using the OHDSI CohortCharacteristics package to generate baseline profiles (“Table 1”) of one or more cohorts in the OMOP CDM, directly addressing the guide’s descriptive design.
How CohortCharacteristics works
- Summarise -> table/plot:
summariseCohortCount/summariseCharacteristicsproduce tidysummarised_resultoutputs consumed bytable…/plot…helpers invisOmopResults. - Cohorts in, summaries out: You provide one or more cohort tables (OMOP CDM structure). Functions compute counts, attrition, demographics, and clinical features at/before index.
- Windows: For granular baselines, large-scale summaries can use windows (e.g., -365:-1, 0:0, 1:365) around index.
- Groups/strata: Each cohort produces groups; you can stratify or facet by
cohort_nameand other variables in plots/tables.
Practical guidance
- Use mock data for reproducibility.
- Keep cohort inclusion/exclusion logic in cohort construction; keep
summarisefunctions descriptive. - Save the
summarised_resultobject and then render tables/plots.
Step 1: Setup
This initial step focuses on preparing the R environment for the analysis. It involves loading libraries like CDMConnector and CohortCharacteristics makes their functions available for data manipulation and analysis. This foundational step is crucial for a reproducible and error-free workflow.
library(CDMConnector)
library(CohortCharacteristics)
library(PatientProfiles)
library(visOmopResults)
library(DrugUtilisation)
Step 2: Create a mock CDM and cohort
To ensure the analysis is reproducible and self-contained, we generate a mock Common Data Model (CDM) using the DrugUtilisation package. This simulated dataset mimics the structure of a real OMOP CDM, providing a safe and standardized environment for developing and testing analytical code. After creating the CDM, we define a cohort of interest—in this case, individuals exposed to acetaminophen. This step is vital for establishing the study population that will be characterized in the subsequent steps.
# Create a mock CDM object for demonstration purposes.
# This simulates a real CDM and populates it with synthetic data.
cdm <- DrugUtilisation::mockDrugUtilisation(numberIndividuals = 10000)
# Generate a cohort of individuals based on the ingredient "acetaminophen".
# This cohort will be used for the characterization analysis.
cdm <- generateIngredientCohortSet(
cdm = cdm,
name = "cohort",
ingredient = "acetaminophen"
)
Step 3: Cohort counts and attrition
Before diving into detailed characterization, it’s important to understand the size and composition of the cohort. This step involves calculating the number of subjects and records in the generated cohort and examining the attrition process, which details how many individuals were excluded at each stage of the cohort definition. These summaries provide a high-level overview of the study population and help identify any potential issues with the cohort definition, ensuring the transparency and validity of the analysis.
# Summarize the number of subjects and records in the cohort.
counts <- summariseCohortCount(cdm$cohort)
# Display the cohort counts in a table.
tableCohortCount(counts)
| CDM name | Variable name | Estimate name | Cohort name |
|---|---|---|---|
| acetaminophen | |||
| DUS MOCK | Number records | N | 7,280 |
| Number subjects | N | 5,975 |
# Summarize the attrition of the cohort, showing how many subjects are dropped at each step of the cohort definition.
attr <- summariseCohortAttrition(cdm$cohort)
# Display the cohort attrition in a table.
tableCohortAttrition(attr)
| Reason | Variable name | |||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| DUS MOCK; acetaminophen | ||||
| Initial qualifying events | 7,297 | 5,975 | 0 | 0 |
| Collapse records separated by 1 or less days | 7,280 | 5,975 | 17 | 0 |
Step 4: Baseline characteristics (“Table 1”) and plots
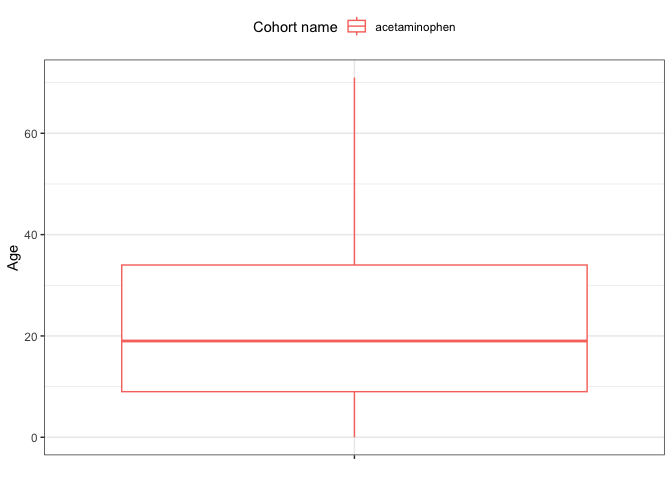
This is the core of the patient-level characterization analysis. We generate a comprehensive summary of the cohort’s baseline characteristics, often referred to as “Table 1.” This table includes demographics, clinical events, and other relevant attributes of the study population at the time of cohort entry. Following the summary, we create visualizations to explore specific aspects of the data, such as the age distribution of the cohort. This step provides deep insights into the study population, which is essential for interpreting the results of any subsequent analyses.
# Summarize the baseline characteristics of the cohort.
# This creates a "Table 1" with demographics, conditions, drugs, etc.
chars <- summariseCharacteristics(cdm$cohort)
# Display the baseline characteristics in a table.
tableCharacteristics(chars)
| CDM name | |||
|---|---|---|---|
| DUS MOCK | |||
| Variable name | Variable level | Estimate name | Cohort name |
| acetaminophen | |||
| Number records | - | N | 7,280 |
| Number subjects | - | N | 5,975 |
| Cohort start date | - | Median [Q25 - Q75] | 2013-03-26 [2002-04-18 - 2019-05-28] |
| Range | 1954-04-29 to 2022-12-31 | ||
| Cohort end date | - | Median [Q25 - Q75] | 2015-08-03 [2006-08-08 - 2020-06-06] |
| Range | 1954-05-17 to 2022-12-31 | ||
| Age | - | Median [Q25 - Q75] | 19 [9 - 34] |
| Mean (SD) | 22.71 (16.73) | ||
| Range | 0 to 71 | ||
| Sex | Female | N (%) | 3,527 (48.45%) |
| Male | N (%) | 3,753 (51.55%) | |
| Prior observation | - | Median [Q25 - Q75] | 663 [171 - 2,049] |
| Mean (SD) | 1,627.62 (2,393.99) | ||
| Range | 0 to 20,955 | ||
| Future observation | - | Median [Q25 - Q75] | 700 [181 - 2,194] |
| Mean (SD) | 1,726.26 (2,519.87) | ||
| Range | 0 to 22,957 | ||
| Days in cohort | - | Median [Q25 - Q75] | 282 [60 - 1,044] |
| Mean (SD) | 937.70 (1,664.07) | ||
| Range | 1 to 18,119 | ||
# Create a plot of the age distribution by cohort.
# First, filter the characteristics data to select only the "Age" variable.
plot_data <- dplyr::filter(chars, variable_name == "Age")
# Generate a boxplot of the age distribution, colored by cohort name.
plotCharacteristics(plot_data, plotType = "boxplot", colour = "cohort_name")
Step 5: Advanced Characterization with Flags
The summariseCharacteristics function offers powerful features for creating custom variables. One such feature is conceptIntersectFlag, which allows you to generate binary flags indicating the presence of specific concepts for each individual in the cohort. This is particularly useful for assessing the prevalence of comorbidities, prior medications, or other specific clinical events. In this step, we’ll create flags for a few example concepts to demonstrate how to enrich the baseline characteristics table.
# Define a list of concepts to create flags for.
# We'll use arbitrary concept IDs for this example,
# representing conditions like 'headache' and 'fever'.
cdm$cohort |>
addSex() |>
summariseCharacteristics(
demographics = FALSE,
strata = list("sex"),
conceptIntersectFlag = list(
"Concomitant disorders" = list(
conceptSet = list(headache = 378253, asthma = 317009, influenza = 4266367),
window = c(-Inf, Inf)
))
) |>
# Display the characteristics table, which now includes the new flags.
tableCharacteristics(
header = c("sex"),
hide = c("cdm_name", "cohort_name", "variable_level", "table_name")
)
| Variable name | Variable level | Estimate name | Sex | ||
|---|---|---|---|---|---|
| overall | Female | Male | |||
| Number records | - | N | 7,280 | 3,527 | 3,753 |
| Number subjects | - | N | 5,975 | 2,904 | 3,071 |
| Concomitant disorders | Headache | N (%) | 3,482 (47.83%) | 1,666 (47.24%) | 1,816 (48.39%) |
| Influenza | N (%) | 3,588 (49.29%) | 1,740 (49.33%) | 1,848 (49.24%) | |
| Asthma | N (%) | 3,563 (48.94%) | 1,719 (48.74%) | 1,844 (49.13%) | |