Population-Level Epidemiology Guide
Introduction & Purpose
Population-Level Epidemiology studies are designed to answer the most fundamental questions in public health: how common is a disease within a population, and is its frequency changing over time? This methodology provides a high-level, panoramic view of a disease’s burden on a community or healthcare system, which is essential for planning public health policies, allocating resources, and identifying emerging health trends.
The purpose is to quantify the frequency of health outcomes at a population level, typically by measuring:
- Incidence: The rate of new cases of a disease in a population over a specific period. This helps us understand the risk of developing the disease.
- Prevalence: The proportion of a population that currently has a disease at a specific point in time (point prevalence) or over a period (period prevalence). This helps us understand the overall burden of the disease.
Study Design
The standard design for this analysis is a population-level cohort study. This involves taking the entire population captured within a database and following them over a defined period of calendar time to observe the occurrence of health outcomes.
Participants
The study typically includes the entire source population available in the database who have a minimum period of data visibility (e.g., at least one year) before the study’s start date. Key considerations for defining the participant group include:
- For Incidence Calculation: To measure only new cases, individuals who have a history of the disease (prevalent cases) are excluded from the analysis. This “washout” period ensures that the cases being counted are genuinely new diagnoses.
- Subpopulations: The analysis can be restricted to specific subpopulations of interest, for example, by limiting the study to people over a certain age or with a specific clinical history.
Outcomes
The primary outcomes are the calculated rates of incidence and prevalence for the disease of interest. These are typically stratified by:
- Demographics: Age groups and gender.
- Time: Calendar year, month, or quarter to observe trends.
Follow-up
The follow-up for the study begins at a pre-defined calendar date (the index date) and continues for a specified period (e.g., one year). This process is often repeated for multiple consecutive periods to generate trend data over several years.
Analyses
The analytical approach is descriptive. The core calculations are:
- Incidence Rate: The number of newly diagnosed people (the numerator) divided by the total person-time at risk in the population (the denominator).
- Prevalence: The number of people with the diagnosis (both new and pre-existing) divided by the total number of people in the source population at a specific point in time or over a period.
How to Implement This Study
The OHDSI IncidencePrevalence R package is designed to calculate population-level incidence and prevalence rates from OMOP CDM data, directly addressing the core analytical requirements of this study design.
At a high level, IncidencePrevalence turns OMOP CDM cohorts into tidy, stratified summaries by repeatedly applying three ideas:
- Denominator time: People contribute at-risk time only when they meet the study rules you set with
generateDenominatorCohortSet()/generateTargetDenominatorCohortSet(). Entry is the latest of study start, prior observation met, and minimum age; exit is the earliest of study end, observation end, and age upper limit. - Outcome overlap: Outcomes are read from a separate cohort table you prepare (here provided by
mockIncidencePrevalence()). Prevalence checks whether a person is in the outcome cohort at a time point/over an interval; incidence counts first qualifying onsets, respecting washout and repeated event rules. - Interval slicing: The study time is cut into calendar intervals (years, quarters, months). Each interval gets its own denominator, outcome counts, and derived measures.
Key computations and arguments
- Point prevalence (
estimatePointPrevalence):- What: proportion with the outcome at a time point inside each interval.
- How: outcome presence on the chosen
timePoint(“start”/“middle”/“end”) among those in the denominator at that date. - Tip: Use when you want a snapshot at a consistent anchor within each interval.
- Period prevalence (
estimatePeriodPrevalence):- What: proportion with the outcome at any time during the interval.
- How: outcome occurs at any day overlapping the interval; denominator includes those present during the interval.
fullContribution: ifTRUE, only people observed for the entire interval count in the denominator; ifFALSE, anyone observed for ≥1 day counts.- Tip: Choose
fullContributionbased on whether incomplete observation could bias proportions for long intervals.
- Incidence (
estimateIncidence):- What: new-onset rates per person-time in each interval.
- How: counts qualifying outcome onsets and divides by person-time contributed in the denominator.
outcomeWashout: requires no outcome in the prior N days; useInfto ensure first-ever incidence.repeatedEvents: ifTRUE, allows multiple incident events per person separated by washout; ifFALSE, at most one incident event per person.completeDatabaseIntervals: ifTRUE, drops intervals where the database cannot establish full observation coverage (e.g., first/last years with partial data), reducing edge bias.
Stratification and groups
- Groups: Each combination of denominator settings (age group, sex, prior history, time-at-risk windows, target-based subsets) becomes a separate
cohort_definition_idand produces separate rows. - Strata: You can facet/colour or summarize by variables such as
denominator_age_group,denominator_sex, etc. UseplotIncidence()/plotPrevalence()helpers andavailable...Grouping()to see valid aesthetics.
Outputs
- Summarised results: Functions return a “summarised_result” tables with
estimate_name/valuepairs, plus additional metadata (start/end dates, interval type). These are directly consumable byvisOmopResultstotableIncidence/tablePrevalenceand plot functions.
Practical guidance
- Pick intervals aligned to your question and data density (years for long-term trends, months for seasonality).
- Use
outcomeWashoutandrepeatedEventsto encode “first-ever” vs. “episode-based” incidence. - Prefer
completeDatabaseIntervals(incidence) and considerfullContribution(period prevalence) when edge effects or intermittent observation could bias estimates. - Keep exclusion logic out of outcome cohorts; define restrictions on the denominator instead.
Our workflow will follow a simple, powerful pattern common across OHDSI packages: summarise -> table/plot.
Step 1: Load Libraries & Connect to the Database
First, we load the necessary libraries. CDMConnector provides the tools to connect to and work with an OMOP CDM database. IncidencePrevalence is the analytical engine we will use to calculate incidence and prevalence rates. Finally, visOmopResults will help us to visualise the results.
For this tutorial, we will use mockIncidencePrevalence() to create a self-contained, reproducible mock database (cdm) object. This allows us to run the analysis without needing a real database connection. The mock data includes a person table, an observation_period table, and an outcome cohort table, which are the minimum requirements for the IncidencePrevalence package.
# Load necessary libraries from the renv library
library(CDMConnector)
library(IncidencePrevalence)
library(visOmopResults)
# Create a mock database connection with sample data
cdm <- mockIncidencePrevalence(sampleSize = 50000)
Step 2: Define the Study Population (The Denominator)
As per our protocol, the study requires a defined source population over a specific time period. We will use generateDenominatorCohortSet() to create our analysis population. This function defines the denominator cohort based on observation period start and end dates, age, and sex. Here, we define our study period from 2008 to 2019 and create strata for two age groups (0-64 and 65-100) and by sex.
# Define the denominator cohort for the study period
cdm <- generateDenominatorCohortSet(
cdm = cdm,
name = "denominator",
cohortDateRange = as.Date(c("2008-01-01", "2019-01-01")),
ageGroup = list(
c(0, 64),
c(65, 100)
),
sex = c("Male", "Female", "Both"),
)
Step 3: Calculate Incidence & Prevalence (The summarise Step)
This step is the core of our analysis, equivalent to running a PROC in SAS. We will use the IncidencePrevalence package to “summarise” the data by calculating incidence and prevalence rates.
Our protocol requires stratifying by time, so we will set interval = "years". We also use the outcomeWashout parameter to ensure we are only counting new cases for our incidence calculation, a key requirement of the protocol.
3.1: Estimate Incidence
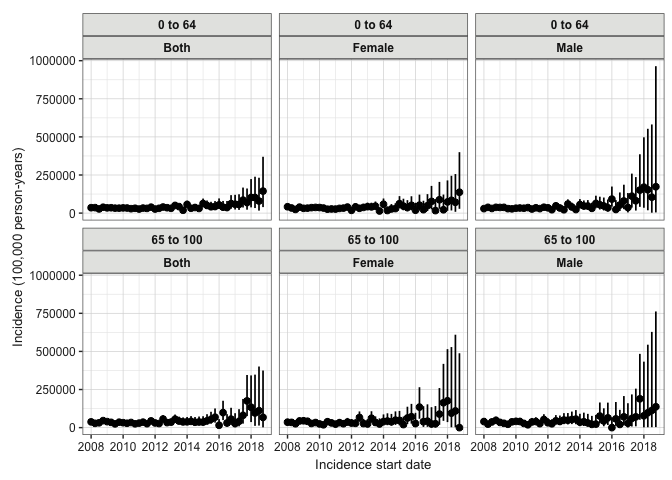
Now we will calculate the incidence of our outcome. We use estimateIncidence() for this. We specify our denominator and outcome tables. We set interval to “quarters” to get quarterly estimates. outcomeWashout is set to Inf to only include the first occurrence of the outcome for each person. repeatedEvents is FALSE, meaning a person cannot have the outcome more than once. Finally, we use plotIncidence to visualise the results, faceting by age group and sex.
# Calculate incidence rates, stratified by year
inc <- estimateIncidence(
cdm = cdm,
denominatorTable = "denominator",
outcomeTable = "outcome",
interval = "quarters",
outcomeWashout = Inf, # Require no prior history of the outcome
repeatedEvents = FALSE
)
# View the summarised results
plotIncidence(inc, facet = c("denominator_age_group", "denominator_sex"))
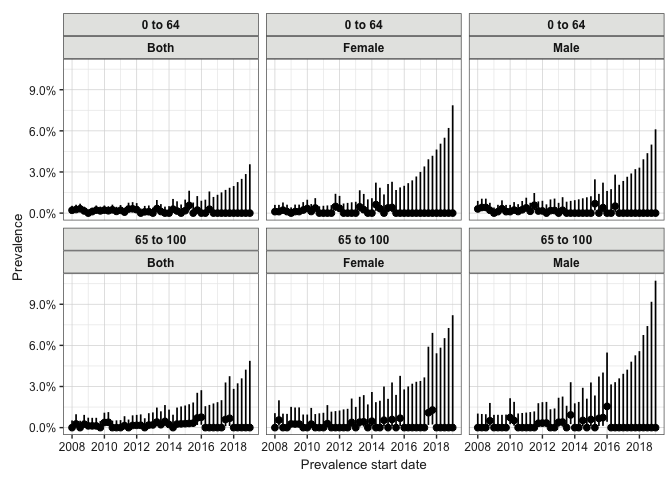
3.2: Estimate Prevalence
Next, we’ll estimate the point prevalence. We use estimatePointPrevalence(), specifying our denominator and outcome tables. We again set the interval to “quarters”. The timePoint parameter is set to “start”, meaning prevalence will be calculated at the start of each quarter. We then use plotPrevalence to visualise the results, again faceting by age group and sex.
# Calculate point prevalence, stratified by year
prev_point <- estimatePointPrevalence(
cdm = cdm,
denominatorTable = "denominator",
outcomeTable = "outcome",
interval = "quarters",
timePoint = "start"
)
plotPrevalence(prev_point, facet = c("denominator_age_group", "denominator_sex"))